Principle
SOD is a metalloenzyme widely present in organisms and is an important oxygen radical scavenger that catalyzes the divergence of superoxide anions to generate H2O2 and O2.SOD is not only a superoxide anion scavenging enzyme, but also a major H2O2-generating enzyme, which plays an important role in the biological antioxidant system.
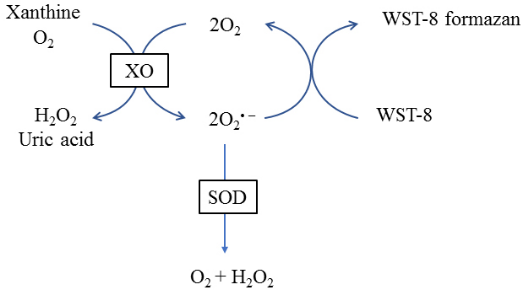
Through the reaction system of xanthine and xanthine oxidase to produce superoxide anion ( O2- ), and WST-8 reaction to generate water-soluble purple dirty; SOD can scavenge O2-, thus inhibiting the formation of dirty; the deeper the purple color of the reaction solution, the lower the activity of SOD, and vice versa, the higher the activity.
Appliance
Detection of SOD activity in samples.
Operation method
Determination of superoxide dismutase (SOD) activity in Cryptobacterium hirsutum nematodes
Principle
SOD is a metalloenzyme widely found in organisms and is an important oxygen radical scavenger that catalyzes the divergence of superoxide anions to generate H2O2 and O2.SOD is not only a superoxide anion scavenging enzyme, but also a major H2O2-generating enzyme, which plays an important role in the biological antioxidant system. Through the xanthine and xanthine oxidase reaction system to produce superoxide anion (O2-), and WST-8 reaction to generate water-soluble purple dirty; SOD can scavenging O2-, thus inhibiting the formation of dirty; reaction solution, the deeper the purple color, that is, the lower the SOD activity, and vice versa, the higher the activity.
Materials and Instruments
SOD Assay Kit, 60 mm Petri Dish Move A. Sample Preparation 1. Sample collection L4 stage nematodes were used as a reference point throughout the protocol. L4 nematodes from each nematode growth plate were washed into a 1.5 mL centrifuge tube with 1.6 mL of phosphate buffered saline (PBS, pH 7.0). After 30 min of gravity settling at 4 °C, the supernatant was carefully removed and discarded. Add 500 μL of pre-cooled PBS, gently resuspend the precipitate, and further settle for 15 min. Then, discard the supernatant, add 500 μL of pre-cooled PBS, gently resuspend, and obtain populations of 20, 50, 100, 200, 300, 400, and 500 nematodes in centrifuge tubes with 200 μL of PBS according to the following procedure. A population of 100 nematodes is used as an example: a. Count the nematodes in 20 μL of PBS under a microscope in 3 separate independent trials to estimate the density of nematodes in the entire tube. b. Based on the number of nematodes obtained, calculate and adjust the volume of PBS so that the final density is 9~11 nematodes per 20 μL. For example, if the number is greater than 10, add more PBS accordingly; if the number is less than 10, perform a 10 min settling and remove the appropriate amount of PBS to achieve the desired density. c. Transfer 260 μL of PBS with nematodes to a new 1.5 mL tube using a pipette and gently repeat to resuspend the sediment. d. Count the number of nematodes in 20 μL of PBS in three independent trials, with gentle repetitive pipetting in between to confirm nematode density, leaving 200 μL in the tube. If nematode density does not match the setup, repeat step A1b. Prepare samples with other nematodes in the same way. 2. Sample Storage Centrifuge the sample at 5000 x g for 5 min (4 °C). Carefully remove the supernatant with a pipette and store the precipitate at -20 °C (overnight) or -80 °C (for more than one week). 3. Sample Homogenization a. Grind the pellet in the centrifuge tube on ice. b. Centrifuge at 5000 x g for 5 min at 4 °C. Aspirate 200 μL of supernatant. Dispense 50 μL into four tubes for subsequent determination.
1.5 mL Tube
1. Prepare standards, controls and working solutions according to the ELISA kit instructions.
a. For each sample, 160 μL of WST-8/Enzyme Working Solution and 20 μL of Reaction Solution are required. Calculate the total amount based on the number of nematode samples and the number of standards. For WST-8/Enzyme Working Solution, 160 μL consists of 151 μL of SOD Assay Buffer, 8 μL of WST-8, and 1 μL of Enzyme Solution. b. Combine the stock solution 1:40 with the standard solution.
b. Dilute the stock solution 1:40 into the SOD Assay Buffer (i.e., 1 μL stock solution to 39 μL buffer) to prepare the reaction solution.
2. Sample Determination
a. Add 20 μL of standard and control to the 96-well plate, and set up at least 2 duplicate wells.
b. Add 20 μL of nematode samples to the 96-well plate and set up at least 2 duplicate wells. It is important to note that after the sample homogenization step, each nematode sample is processed and divided into 4 portions to avoid repeated freezing and thawing of the samples.
c. Using a multichannel pipette, add 160 μL of WST-8/enzyme working solution to each sample, followed by 20 μL of reaction solution. The standard concentrations were 100, 50, 10, 5, 2.5, 1.25, and 0.625 U/mL.
d. There were two blank controls. Blank Control 1 was made by replacing 20 μL of sample with SOD Assay Buffer. Blank Control 2: Replace 20 μL of sample and 20 μL of reaction solution with 40 μL of SOD Assay Buffer.
e. Incubate at 37 ℃ for 30 min.
f. Read the absorbance at 450 nm (A450) with an enzyme meter.
3. Calculation of SOD activity
a. Inhibition (%) = (Blank Control 1 - Standard or Sample) / (Blank Control 1 - Blank Control 2) x 100%. A standard curve was established using a standard with known SOD activity and its inhibition. The curve is then used to calculate the SOD activity of the samples.
b. The total protein concentration of each sample is used as an internal reference to eliminate differences in nematode populations between samples, and the standardized SOD activity is expressed as the ratio of the measured SOD activity to the total protein concentration of that sample.
The table below lists the standardized SOD activities in nematodes, with the ratios generally ranging from 4.04 to 4.35, however the values for 20 and 50 nematodes/group are much larger than the general range. Therefore a minimum of 100 nematodes/group is required to ensure the feasibility and stability of SOD activity measurements.

4. Data Analysis
a. At least 3 aliquots of each sample, with at least 3 assay wells per aliquot, are analyzed for total nematode protein (TP) and the mean TP value for each aliquot is calculated to represent the TP concentration in the sample. b. The SOD value for each well is calculated for each nematode. c. The SOD value for each aliquot is calculated for each nematode. Calculate the ratio of the SOD value to the mean TP value for each well to obtain the SOD activity in the sample.
b. If the samples are from different treatments, normalize the normalized SOD activity in the control to 100% (or 1.0); the normalized SOD activity in the treatment group is then converted to percent of controls (POCs) (or fold-change of controls); the average POC is then calculated using the POC value (or fold-change) in the sample.
Caveat
1. When nematodes are collected from growth plates, bacteria in the worm slurry can interfere with the assay. Therefore, instead of centrifugation, gravity settling is used for sample collection. To completely remove the bacteria, gently wash the sediment (i.e. resuspend - settle - discard supernatant) several times.
2. to ensure reproducibility, there should be at least 100 nematodes in each tube.
3. the supernatant of a homogenized PBS solution from 100 nematodes is sufficient for four aliquots, which is sufficient for four biochemical assays including total protein concentration. If more biochemical assays are performed simultaneously, the number of nematodes should be increased.
For more product details, please visit Aladdin Scientific website.
